ChemX: A Collection of Chemistry Datasets for Benchmarking Automated Information Extraction¶
A multimodal benchmark for evaluating machine learning models that extract structured chemical data from scientific literature.
About the Project¶
ChemX is a curated benchmark suite aimed at assessing and improving the performance of AI systems in extracting structured chemical information from scientific articles across multiple modalities: text, tables, and figures.
The benchmark covers diverse chemical topics, including nanomaterials, small molecules, chelate complexes, and their properties relevant for various applications.
Project Goal
Enable reliable and scalable chemical knowledge extraction by combining multimodal data annotation with expert validation, thereby accelerating downstream scientific research.
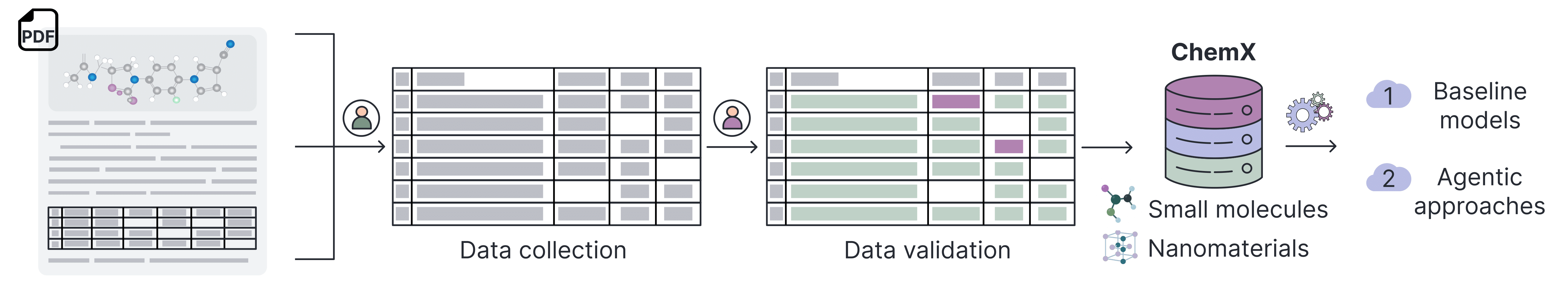
Key Features¶
- 10 manually annotated datasets across a range of chemical subfields
- Over 16,000 structured records extracted from peer-reviewed literature
- Multimodal data sources: text passages, tables, chemical diagrams, figures
- Provenance and annotation metadata for every data point
- Expert-reviewed annotations ensuring high data quality
- Standardized evaluation benchmarks for NLP, vision, and multimodal models
Why It Matters¶
Despite advances in large language and vision-language models, scientific chemistry lags behind in AI adoption due to the lack of reliable, multimodal, annotated benchmarks.
The Solution
ChemX closes this gap by providing a transparent, high-quality benchmark for evaluating and training data extraction systems, validated by domain experts.
How to Use ChemX¶
- Train and test models to extract chemical entities, values, units, and relationships
- Compare performance across extraction tasks using a unified evaluation framework
- Explore multimodal learning for chemistry-specific document analysis
- Use structured chemical data for tasks like toxicity modeling, material design, and reaction planning
Site Sections¶
Overview¶
Motivation, annotation pipeline, and an overview of all datasets.
Datasets¶
Nanomaterials¶
- Cytotoxicity Dataset — Nanoparticle toxicity in mammalian cells
- SelTox Dataset — Nanoparticle toxicity in microbial systems
- Synergy Dataset — Antibiotic–nanoparticle interaction effects
- Nanozymes Dataset — Enzymatic activity of nanozymes
- Magnetic Nanomaterials — Magnetic property extraction
Small Molecules¶
- Benzimidazole Antibiotics — Inhibitory concentrations of benzimidazoles
- Oxazolidinone Antibiotics — Activity profiling of oxazolidinones
- Chelate Metal Complexes — Thermodynamic parameters of chelates
- Eye Drops — Corneal permeability and pharmaceutical properties
- Cocrystal Photostability — Stability of pharmaceutical co-crystals
Methods¶
Detailed pipeline for annotation, validation, and benchmarking:
Guideline¶
Examples for training and evaluating extraction models with CDEB datasets.
About the Project¶
Team, how to cite the project, version history, and acknowledgments.